From structure factors to density using Fourier synthesis
fousynth.RdGiven a set of structure factors, separately as a vector of amplitudes and a vector of phases in degrees, corresponding to a set of 1D Miller indices, the length of the 1D unit cell, the set of Miller indices and the number of grid points used to calculate the 1D density, this function calculates the 1D density corresponding to the given structure factors.
Arguments
- a
A real number. The unit cell side length.
- Fmod
A vector of real numbers. The structure factors' amplitudes corresponding to the 1D density to be calculated.
- Fpha
A vector of real numbers. The structure factors' phases (in degrees) corresponding to the 1D density to be calculated.
- hidx
A vector of integer numbers. The set of 1D Miller indices corresponding to the set of structure factors F.
- N
An integer number. The number of grid points used to calculate the 1D density.
Value
A vector of N real numbers representing the calculated 1D density at each of the regular N grid points.
Examples
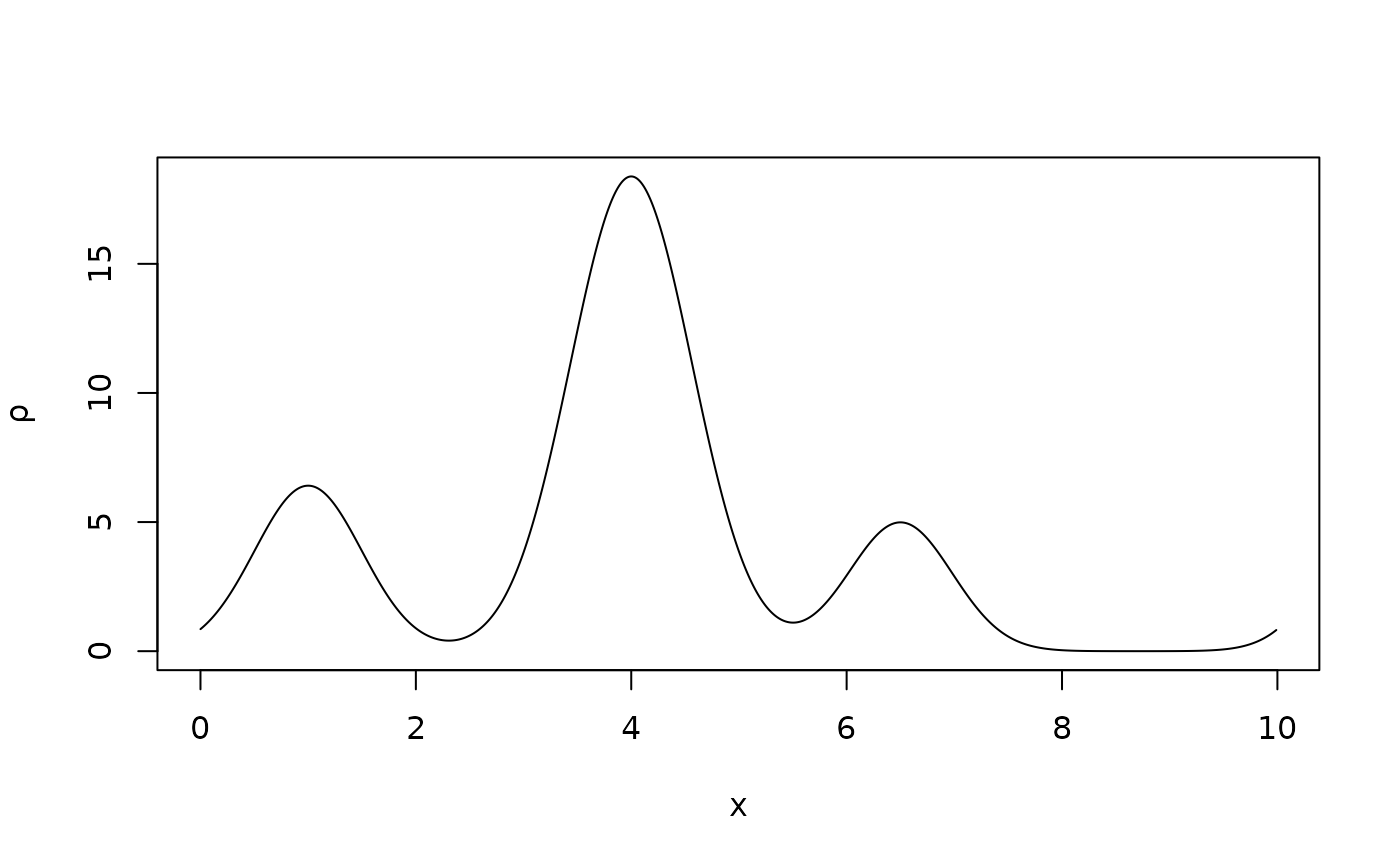
# First create the crystal structure (in P1)
a <- 10
SG <- "P1"
x0 <- c(1,4,6.5)
Z <- c(8,26,6)
B <- c(18,20,17)
occ <- c(1,1,1)
sdata <- standardise_sdata(a,SG,x0,Z,B,occ)
# Enough Fourier components (Miller indices)
hidx <- 0:20
# Compute the structure factors
ftmp <- strufac(hidx,sdata)
# Number of grid points
N <- 1000
# Density
rtmp <- fousynth(a,ftmp$Fmod,ftmp$Fpha,hidx,N)
# Density plot in the unit cell
x <- rtmp$x
rho <- rtmp$rr
plot(x,rho,type="l",xlab="x",ylab=expression(rho))